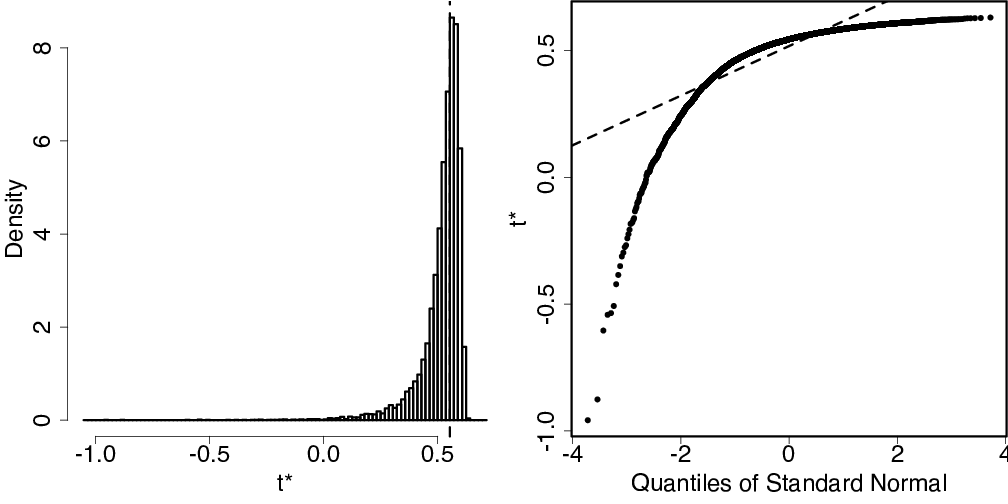
R code used to produce the graph shown below.
result <- boot(mydata, heritability, R=10000)
result
plot(result)
plotmyboot <- function (x, index = 1, t0 = NULL, t = NULL, jack = FALSE, qdist = "norm",
nclass = NULL, df, ...)
{
boot.out <- x
t.o <- t
if (is.null(t)) {
t <- boot.out$t[, index]
if (is.null(t0))
t0 <- boot.out$t0[index]
}
t <- t[is.finite(t)]
if (boot:::const(t, min(1e-08, mean(t, na.rm = TRUE)/1e+06))) {
print(paste("All values of t* are equal to ", mean(t,
na.rm = TRUE)))
return(invisible(boot.out))
}
if (is.null(nclass))
nclass <- min(max(ceiling(length(t)/25), 10), 100)
if (!is.null(t0)) {
rg <- range(t)
if (t0 < rg[1L])
rg[1L] <- t0
else if (t0 > rg[2L])
rg[2L] <- t0
rg <- rg + 0.05 * c(-1, 1) * diff(rg)
lc <- diff(rg)/(nclass - 2)
n1 <- ceiling((t0 - rg[1L])/lc)
n2 <- ceiling((rg[2L] - t0)/lc)
bks <- t0 + (-n1:n2) * lc
}
R <- boot.out$R
if (qdist == "chisq") {
qq <- qchisq((seq_len(R))/(R + 1), df = df)
qlab <- paste("Quantiles of Chi-squared(", df, ")", sep = "")
}
else {
if (qdist != "norm")
warning(gettextf("%s distribution not supported: using normal instead",
sQuote(qdist)), domain = NA)
qq <- qnorm((seq_len(R))/(R + 1))
qlab <- "Quantiles of Standard Normal"
}
if (jack) {
layout(mat = matrix(c(1, 2, 3, 3), 2L, 2L, byrow = TRUE))
if (is.null(t0))
hist(t, nclass = nclass, probability = TRUE, xlab = "t*", main="")
else hist(t, breaks = bks, probability = TRUE, xlab = "t*", main="")
if (!is.null(t0))
abline(v = t0, lty = 2)
qqplot(qq, t, xlab = qlab, ylab = "t*")
if (qdist == "norm")
abline(mean(t), sqrt(var(t)), lty = 2)
else abline(0, 1, lty = 2)
jack.after.boot(boot.out, index = index, t = t.o, ...)
}
else {
par(mfrow = c(1, 2))
if (is.null(t0))
hist(t, nclass = nclass, probability = TRUE, xlab = "t*", main="")
else hist(t, breaks = bks, probability = TRUE, xlab = "t*", main="")
if (!is.null(t0))
abline(v = t0, lty = 2)
qqplot(qq, t, xlab = qlab, ylab = "t*")
if (qdist == "norm")
abline(mean(t), sqrt(var(t)), lty = 2)
else abline(0, 1, lty = 2)
}
par(mfrow = c(1, 1))
invisible(boot.out)
}
plotmyboot(result, main="asdasd")
